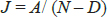International Journal of Agriculture, Environment and Biotechnology
Citation: IJAEB: 12(4): 331-337, December 2019
DOI: 10.30954/0974-1712.12.2019.6
©2019 IJAEB All rights reserved
HORTICULTURE
Molecular Characterization of Different Strawberry (Fragaria x ananassa Duch.) Cultivars Growing under Mid Hill Conditions of Himachal Pradesh
Sonia Kumari*1, Shashi Kumar Sharma2 and Ankush Kumar3
1Department of Fruit Science, Dr. YS Parmar UHF Nauni, Solan, Himachal Pradesh, India
2Management Expert- Horticulture HP-SHIVA, Directorate of Horticulture, Navbahar Shimla, Himachal Pradesh, India
3National Dairy Research Institute, Karnal, Dairy Economics Division, Haryana, India
*Corresponding author: soniasharma75059@gmail.com (ORCID ID: 0000-0002-8222-6294)
Paper No. 801
Received: 18-06-2019
Revised: 21-10-2019
Accepted: 28-11-2019
ABSTRACT
Genetic similarity of fifteen strawberry cultivars was assessed using RAPD and ISSR molecular markers. The DNA of the fifteen strawberry cultivars were extracted and amplified by PCR with RAPD and ISSR primers. The DNA Fragments were separated in agarose gel for RAPD and in polyacrylamide gel for ISSR markers. The genetic similarity matrix was estimated by Jaccard coefficient. Studied cultivars were found to be coherent into two groups. RAPD markers projected more coherence with origin and genealogy of the cultivars. ISSR markers projected more similarity between ‘Chandler’ and ‘Shasta’ whereas as per RAPD marker ‘Chandler’ and ‘Sweet Charlie’ were more closely related. In molecular assessment Elyana cultivar was found to be marked cultivars as it shows long flowering duration, higher yield and bigger fruits and can be used as a potential genotype to breed cultivars among the locally adapted cultivars.
Highlights
 The present study involves the Molecular Characterization using RAPD and ISSR markers to assess the genetic similarity of fifteen strawberry cultivars.
The present study involves the Molecular Characterization using RAPD and ISSR markers to assess the genetic similarity of fifteen strawberry cultivars.
 ‘Elyana’ cultivar was found to be distinct from other cultivars based on molecular assessment because of its long flowering duration, higher yield and bigger fruits therefore, may be considered as a potential genotype.
‘Elyana’ cultivar was found to be distinct from other cultivars based on molecular assessment because of its long flowering duration, higher yield and bigger fruits therefore, may be considered as a potential genotype.
Keywords: Fragaria x ananassa Duch., Genetic Variability, Jaccard’s Coefficient, Dendrogram, Informative Primers, Polymorphic Bands
The cultivated strawberry (Fragaria x annanasa Duch.) is an octaploid (2n=8x=56) species of family Rosaceae that has economic importance related to fruit yield. The strawberry plant has experienced a series of polyploidization and natural hybridization events in its evolutionary process (Hancok 1996) therefore it is difficult to analyse its genetic base. Most of studies have mainly focused on the morphological, cytological, palynological characteristics and molecular biology research to estimate the genetic diversity and relationship among strawberry resources. Kuras and Korbin (2004) utilized two Polymerase Chain Reaction (PCR)-based techniques, Random Amplified Polymorphic DNA and Inter Simple Sequence Repeats (ISSR), to determine the genetic relationship and germplasm diversity of strawberry cultivars.
Traditionally, cultivar(s) identification of strawberry has been based on morphological traits of leaf, flower and fruit (Dale 1996). This approach is limited in its usefulness, since closely related cultivars are often difficult to distinguish by morphological indices alone. Furthermore, expression of morphological features generally got affected by the environment, including the method of cultivation. Inbreeding in cultivated strawberry has lead to rapid loss of vigour, yield, and fruit size (Morrow and Darrow 1952; Spangelo 1971). The germplasm characterization by DNA based molecular markers allows better understanding of genetic variability. This is due to determination of the level of genetic divergence and the molecular pattern of each cultivar. Several techniques based on DNA analysis are available for genetic divergence studies. In strawberry, Random Amplification of Polymorphic DNA (RAPD) and Inter Simple Sequence Repeats (ISSR) markers are efficient for genetic divergence studies (Gidoni 1994; Hussein 2008). The ISSR technique is a potentially useful tool for the identification of strawberry varieties as it is simple, cost-effective, fast and highly discriminant and reliable (Arnau 2003). The objective of present study is characterization of strawberry cultivars using molecular markers.
MATERIALS AND METHODS
The present investigation was carried out in the Department of Fruit Science, Dr. YS Parmar University of Horticulture & Forestry, Nauni, Solan (HP). The details of experiment conducted, material used & techniques employed for studies have been described briefly.
DNA extraction: The presence of undesirable compounds such as polyphenols and polysaccharides are the two biggest problems faced during the isolation and purification of high quality DNA in plants (Nunes et al., 2011). The isolation of genomic DNA from plants that contain high level of polysaccharides, phenolics, terpenoids, tannins and metabolites is cumbersome because these compounds binds with DNA and get co-extracted with DNA (Micheils et al. 2003 and Puchooa 2004). Isolation of plant DNA (CTAB method) from leaf samples were done as per the technique outlined by Doyle and Doyle (1990).
PCR amplification: The introduction of PCR-based methods constituted a new milestone in the field of DNA fingerprinting. The breakthrough came when a thermostable DNA polymerase, Taq polymerase, was isolated and purified. This allowed the cycling process to be automated as only a single addition of enzyme was required. This way the DNA fragments are enzymatically amplified between pairs of DNA primers that hybridize to the sample DNA. A large number of factors can influence the success of the reaction and the nature of PCR products. Among these, concentration and quality of template DNA, concentration and stability of DNA polymerase, concentration of magnesium ions, temperature profile and primer size are important.
Gel Electrophoresis: Twenty-seven ISSR and 22 RAPD primers were used to carry out DNA amplification of fifteen strawberry cultivars. Data was recorded based on agarose gel for both i.e. ISSR and RAPD profile. PCR products were separated on 1.2% agarose gel which was prepared by dissolving 2.4g of agarose in 200 ml of 10X TAE buffer (for RAPD) and 1.6% agarose gel was prepared by dissolving 3.2 g of agarose in 200 ml of 10X TAE buffer (for ISSR) and followed by heating until solution becomes clear. Solution was cooled to 50-60oC followed by addition of ethidium bromide, mixed thoroughly and poured in gel casting tray and allowed the gel to set for 1 hour and remove the comb carefully. Running tray was transferred to running platform of electrophoresis unit having tank buffer, which was prepared by the addition of 20 ml of 10X TAE (for RAPD) and 20 ml of 50X TAE (for ISSR) in 980 ml of distilled water to make final volume 1000 ml. DNA sample were prepared by mixing PCR products and 3µl of 10X loading dye. After cleaning wells carefully with running buffer DNA samples were loaded with DNA sized marker. Electrophoresis was carried out at 80 V for 3-4 hours. Gel was viewed under U.V. transilluminator and photographed using Gel Documentation System (Bio Rad).
Data Analysis: Most of the primers of ISSR were polymorphic while some of them did not amplify, whereas in RAPD very less number of primers gave polymorphic bands out of which few gave smeared bands while few of them produced monomorphic bands however they are not included in data analysis. The presence of band is represented by 1 and absence of band by 0. Data analysis was carried out only for those primers which give scorable patterns for the genotypes under study. Co- migrating bands were considered to represent the same locus and thus treated as a same band while scoring. Intensity of the amplified products was not taken into account while scoring. NTSYS-pc, version 2.02 or version 1.8 (Numerical Taxonomy and Multivariate Analysis System) was used to perform cluster analysis of the complete RAPD data. Similarity between accession were estimated using the Jaccard coefficient, calculated as:
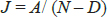
Where,
A is the no. of positive matches (i.e. the presence of bands in both the samples), D is the no. of negative matches (i.e. the absence of bands in both the samples), N is the total sample size including the no. of matches and unmatches. Similarity estimates were analysed by Un-weighted Pair Group Method with Arithmetic Averages (UPGMA) and the resulting clusters were expressed as dendrogram in the strawberry genotypes.
RESULTS AND DISCUSSION
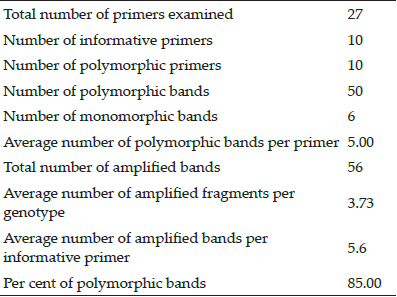
Only 10 out of 27 ISSR primers used were able to produce amplification on PCR analysis and hence were called informative. Among the fifty-six bands produced, fifty were found to be polymorphic, representing 85.00 per cent polymorphism. Two bands were found to be unique and average number of polymorphic bands per primer was recorded to be 5.00 and average number of amplified fragments per cultivar was 3.73 (Table 1). Average number of amplified fragments per informative primer was found 5.6. Present results are in line with the work done by Morales (2011) who also reported 100 per cent polymorphism with primers namely ISSR 827, ISSR 845, ISSR 848 and ISSR 860 in different strawberry cultivars.
Table 1: Summary of amplified products obtained in 15 Strawberry genotypes using ISSR primers
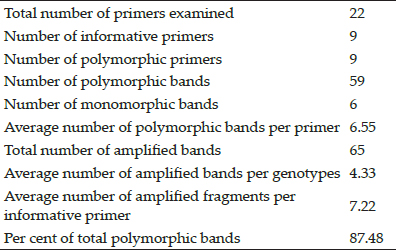
At the same time only 9 out of 22 RAPD primers used were able to produce amplification on PCR analysis and hence were called informative. Among the 65 bands produced, 59 were polymorphic, representing 87.48 per cent polymorphism. Five bands were marked as unique and average number of polymorphic bands per primer was recorded to be 6.55 and average number of amplified fragments per cultivar was 4.33 as given in table 2. Average number of amplified fragments per informative primer was 7.22.
Table 2: Summary of amplified products in15 strawberry genotypes using RAPD primers
PCR-based molecular markers are playing an increasingly important role in the analysis of genetic diversity of horticulture and field-crop species (Harrison et al. 2000). The similarity values based on ISSR data were reported to be higher than those based on RAPD. In this regard, parallel study using RAPD and ISSR techniques showed that RAPD required the testing of six times more primers than ISSR (Korbin 2002 & Conti et al. 2002).
Table 3: Primer sequence and number of loci amplified and polymorphic in 15 Strawberry Cultivars
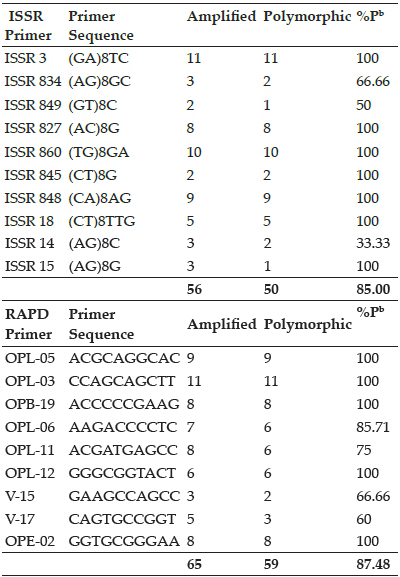
Similarity Coefficient (Jaccard’s Coefficient) Analysis
NTSYSpc ver. 2.02.h (Rohlf 2000) was used to obtain Jaccard’s similarity coefficient matrix (Nei and Li 1979), based on DNA amplification using ISSRs and RAPDs. Good range of genetic variability was found between cultivars as the values of coefficient of variation ranged from 0.26 to 0.77.
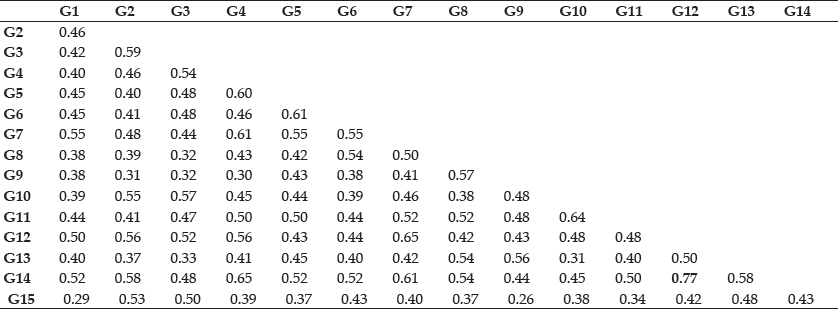
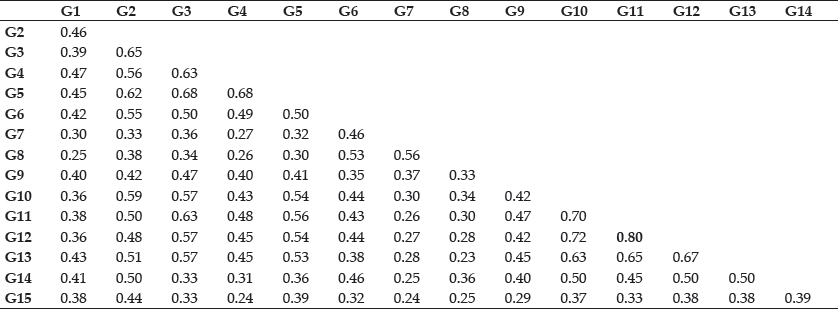
Average of highest and lowest coefficient (Table 4) comes out to be 0.51. The highest value of similarity was 0.77, between G12 (Chandler) and G14 (Shasta). Minimum similarity of 0.25 were obtained between G9 (Torrey) and G15 (Belrubi).
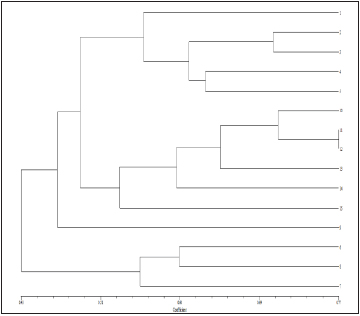
G1. Elyana G2. Gorella G3. Brighton G4. Fern G5. Selva G6.Tioga G7. Confectura G8. Etna G9. Torrey G10. Douglas G11. Sweet Charlie G12. Chandler G13. Addie G14. Shasta G15. Belrubi
Fig. 1: Dendrogram of genetic similarity among fifteen strawberry cultivars, obtained from ISSR markers using the UPGMA method
These values showed high polymorphism and are in close conformity with the investigation as observed by Graham 1996, Arnau 2000 and Kuras 2004. These results are congruent with those of Arnau (2002), who showed the highly discriminatory nature of the ISSR markers.
Results of Hussein (2008) are in disagreement with the present work, who observed efficiency of a few ISSR markers in correctly in identifying and discriminating among strawberry genotypes. This contradiction in results could be due to different cultivars taken for study, lesser number of primers used etc. Jaccard’s similarity coefficient matrix (Nei and Li 1979), based on DNA amplification using 35 ISSR markers, obtained through NTSYSpc ver. 2.02.h (Rohlf 2000) is presented in Table 3. The coefficient values ranged from 0.23 to 0.80 showed a good range of genetic variability in the collections under study. The highest value of similarity was 0.80, between G11 (Sweet Charlie) and G12 (Chandler). Minimum similarity of 0.23 was obtained between G8 (Etna) and G13 (Addie). The present results are in close line with work done by Korbin (2002) and Anita (2004) who also observed lower similarities by RAPD compared to ISSRs marker. This fact is related to low reaptability of RAPD markers caused by its elevated sensitivity to experimental conditions (Jones 1997) which could be the main limiting factor of the technique.
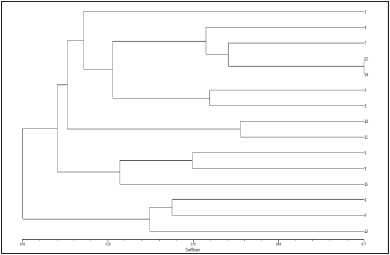
G1. Elyana G2. Gorella G3. Brighton G4. Fern G5. Selva G6.Tioga G7. Confectura G8. Etna G9. Torrey G10. Douglas G11. Sweet Charlie G12. Chandler G13. Addie G14. Shasta G15. Belrubi
Fig. 2: Dendrogram of genetic similarity among fifteen strawberry cultivars, obtained from RAPD markers using the UPGMA method
Cluster Analysis: Dendrogram showing strawberry genotypes into different clusters was based on NTSYSpc ver. 2.02h. Similarity coefficient obtained by Jaccard’s coefficient was further used for UPGMA analysis of NTSYS. SAHN module of NTSYS used the pair wise obtained coefficient to construct dendrograms.
Table 4: Genetic similarity indices between each pair of the fifteen strawberry cultivars (Fragaria x ananassa Duch.) based on ISSR fragment analysis
G1. Elyana G2. Gorella G3. Brighton G4. Fern G5. Selva G6.Tioga G7. Confectura G8. Etna G9. Torrey G10. Douglas G11. Sweet Charlie G12. Chandler G13. Addie G14. Shasta G15. Belrubi
Table 5: Genetic similarity indices between each pair of the fifteen strawberry cultivars (Fragaria x ananassa Duch.) based on RAPD fragments
G1. Elyana G2. Gorella G3. Brighton G4. Fern G5. Selva G6.Tioga G7. Confectura G8. Etna G9. Torrey G10. Douglas G11. Sweet Charlie G12. Chandler G13. Addie G14. Shasta G15. Belrubi
Based on ISSR profile: In the dendrogram, the 15 strawberry were separated into two main clusters, ‘A’ and ‘B’ at 41 per cent similarity value. Cluster A contain 12 genotypes namely, ‘Elyana’, ‘Fern’, ‘Confectura’, ‘Chandler’, ‘Shasta’, ‘Selva’, ‘Tioga’, ‘Douglas’, ‘Sweet Charlie’, ‘Gorella’, ‘Brighton’and ‘Belrubi’ whereas cluster B contained three genotypes viz. ‘Etna’, ‘Torrey’, and ‘Addie’.
Cluster A was further divided into sub clusters i.e. A1 and A2 at similarity value of 43 per cent. Cluster A1 included nine cultivars whereas the other three cultivars were grouped in A2. Cluster A2 was further subdivided into clusters ‘A21’ and ‘A22’ at 45 per cent similarity. Cluster A21 and A22 were further divided into sub clusters at 47 per cent similarity. Maximum similarity of 77 per cent was found between ‘Chandler’ and ‘Shasta’ showing that these two are relatively closely related.
Based on RAPD profile: Fifteen strawberry cultivars were divided into two main clusters, ‘C’ and ‘D’ at 43 % similarity. Cluster C was found to contain twelve genotypes viz and cluster D contained three cultivars viz. ‘Tioga’, ‘Etna’ and ‘Confectura’. Cluster C and D were further subdivided into clusters C21, C22 and D21, D22 at 47 per cent and 55 per cent similarity respectively. Cluster C21 contained eleven genotypes and C22 contained only one genotype. Cluster D21contained two genotype and cluster D22 contained only one genotypes. Maximum similarity i.e. 77 per cent was found between ‘Sweet Charlie’ and ‘Chandler’ showed them to be relatively closely related.
Among diversity analysis, polymorphism with two marker systems showed that these two markers are suitable tools for genetic diversity studies ISSRs and RAPD produced polymorphism which separated fifteen cultivars in two main clusters. Present results based on cluster analysis are in contradiction to that of conclusion drawn by Molares (2011), Kuras (2002) and Korbin (2004) who found similar results with both RAPD and ISSR in strawberry cultivars identification. The variation in results obtained may be due to lesser number of primers used in the presnt study and different cultivars used for evaluation.
CONCLUSION
From the above discussed results it has been concluded that ISSR and RAPD markers revealed variable results. ISSR marker showed that ‘Chandler’ and ‘Shasta’ are more closely related whereas RAPD marker indicated genetic similarity between ‘Chandler’ and ‘Sweet Charlie’. ‘Elyana’ cultivar was found to be distinct from other cultivars based on molecular assessment because of its long flowering duration, higher yield and bigger fruits therefore, may be considered as a potential genotype to breed cultivars among the locally adapted cultivars.
ACKNOWLEDGEMENTS
The authors are thankful to Dr Girish Sharma for providing laboratory facility and experimental supports in the Department of Fruit Science, Dr YS Parmar UHF Nauni, Solan HP. Helps from Dr. Shashi Kumar Sharma and Ankush Kumar are also acknowledged.
REFRENCES
Arnau, G., Lallemand, J. and Bourgoin, M. 2002. Fast and reliable strawberry cultivar identification using inter simple sequence repeat (ISSR) amplification. Euphytica, 129: 69-79.
Anita, K.K., Malgorzata and Edvard, Z. 2004. Comparison of suitability of RAPD and ISSR techniques for determination of strawberry (Fragaria x ananassa Duch.) relationship. In: Plant Cell. Tissue and Organ Culture Springer Netherlands, 79: 189-193.
Conti, J.H., Minani, K. and Games, L.H. 2002. Estimates of genetic similarity and identification of strawberry cultivars by RAPD analysis. Horticultara Brasileira, 20: 145-152.
Dale, A. 1996. A key and vegetative description of thirty-two common strawberry varieties grown in North America. Advances in Strawberry Research, 15: 1-12.
Doyle, J.J. and Doyle, J.L. 1990. Isolation of plant DNA from fresh tissue. Focus, 12: 13-15.
Gidoni, D., Ram, M., Kunik, T., Zur, M., Izsak, S. and Firon, N. 1994. Strawberry cultivar identification using Randomly Amplified Polymorphic DNA (RAPD) markers. Plant Breeding, 113: 339-342..
Graham, J. and McNicol, R.J. 1996. A comparison of method for the estimation of genetic diversity in strawberry cultivars. Theoretical and Applied Genetics, 93: 402-406.
Hancock, J.F. and Shaw, D.V. 1996. Randomly amplified polymorphic DNA in the cultivated strawberry, Fragaria x ananassa. Journal of American Society Horticulture Science, 119: 862-864.
Hussein, T.S., Tawfik, A.A. and Khalifa, M.A. 2008. Molecular identification and genetic relationships of six strawberry varieties using ISSR markers. International Journal Agriculture Biology, 10: 677-80.
Kuras, A. and Korbin, M. 2010. Pco-A analysis of strawberry germplasm used in evaluation of DNA polymorphism of investigation plants. Journal of Fruit and Ornamental Plant Research, 18: 7-16.
Korbin, M. 2002. Fruit plant germplasm characterization using molecular markers in RAPD and ISSR-PCR. Cellular and Molecular Biology Letters, 7: 785-794.
Kuras, A., Korbin, M. and Zurawicz, E. 2004. Comparison of suitability of RAPD and ISSR techniques for determination of strawberry (Fragaria x ananassa Duch.) relationship. The Netherlands, 79: 189-193.
Michiels, A., Van den Endem W., Tuckerm M., Riet, L.V. and Laere, A.V. 2003. Extraction of highquality genomic DNA from latex-containing plants. Analytical Biochemical, 315: 85-89.
Morales, R.G.F., Resende, J.T.V., Faria, M.V. and Resende, L.V. 2011. Genetic similarity among strawberry cultivars assessed by RAPD and ISSR markers. Science Agriculture, 68: 665- 670.
Morrow, E.B. and Darrow, G.M. 1952. Effects on limited in breeding in strawberries. Proceedings of American Society Horticulture Sciences, 59: 269-276.
Nei, M. and Li, W.H. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of National Academyof Sciences USA, 76: 269–5273.
Nunes, C.F. 2011. The genetic diversity of strawberry strawberry (Fragaria x annanasa Duch.) hybrids based on ISSR markers. Acta Scientiarum, 35: 443-452.
Puchooa, D. 2004. A simple, rapid and efficient method for extraction of genomic DNA from lychee (Litchi chinensis Sonn.). African Journal of Biotechnology, 3: 253-255.
Rohlf, F.J. 2000. NTSYS-PC Numerical Taxonomy and Multivariate Analysis System: Manual Applied of Biostatistics. Exeter Software, Setauket, NY, USA.
Spangelo, LP.S, Hsu S.S., Fejer, S.D. and Watkins, R. 1971. Inbreeding line × tester analysis and potential of inbreeding in strawberry breeding. Canadian Journal of Genetic Cytology, 13: 460-469.
 The present study involves the Molecular Characterization using RAPD and ISSR markers to assess the genetic similarity of fifteen strawberry cultivars.
The present study involves the Molecular Characterization using RAPD and ISSR markers to assess the genetic similarity of fifteen strawberry cultivars. ‘Elyana’ cultivar was found to be distinct from other cultivars based on molecular assessment because of its long flowering duration, higher yield and bigger fruits therefore, may be considered as a potential genotype.
‘Elyana’ cultivar was found to be distinct from other cultivars based on molecular assessment because of its long flowering duration, higher yield and bigger fruits therefore, may be considered as a potential genotype.